We presented an abstract at the International Gap Junction Conference, 2015 in Valparaiso, Chile last week.
Authors were: Sigulinsky, L Crystal., Lauritzen, J. Scott., Rapp, Christopher N., Sessions, Alex M., Emrech, Daniel P., Rapp, Kevin D., Watt, Carl B., Anderson, James R., Jones, Bryan W., and Marc, Robert E.
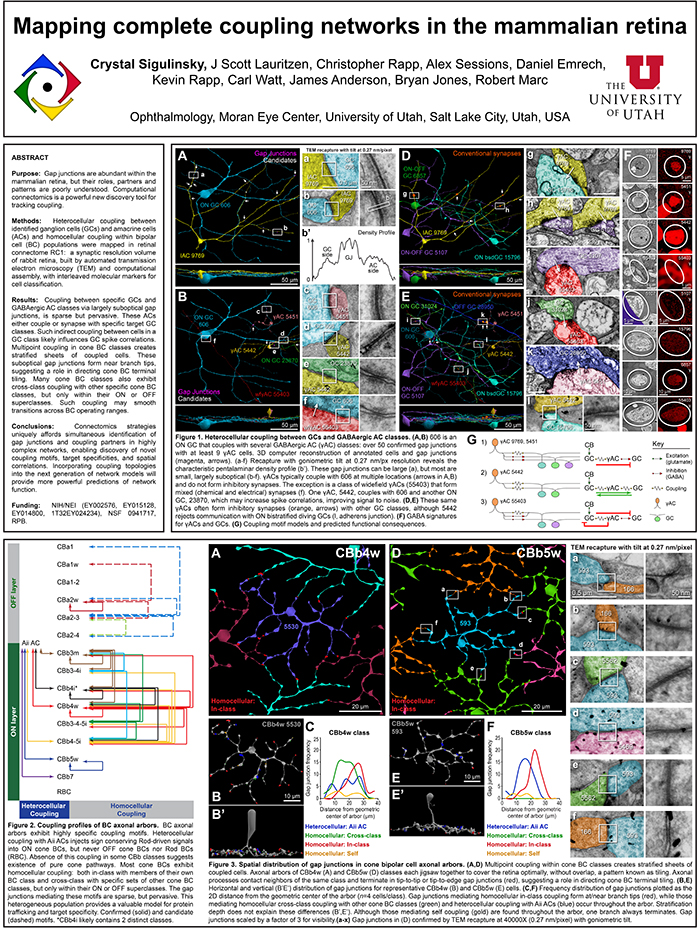
Purpose: Gap junctions are abundant within the mammalian retina, but their roles, partners and patterns are poorly understood. Computational connectomics is a powerful new discovery tool for tracking coupling.
Methods: Heterocellular coupling between identified ganglion cells (GCs) and amacrine cells (ACs) and homocellular coupling within bipolar cell (BC) populations were mapped in retinal connectome RC1: a synaptic resolution volume of rabbit retina, built by automated transmission electron microscopy and computational assembly, with interleaved molecular markers for cell classification.
Results: Coupling between specific GCs and GABAergic AC classes via largely suboptical gap junctions, is sparse but pervasive. These ACs either couple or synapse with specific target GC classes. Such indirect coupling between cells in a GC class likely influences GC spike correlations. Multipoint coupling in cone BC classes creates stratified sheets of coupled cells. These suboptical gap junctions form near branch tips, suggesting a role in directing cone BC terminal tiling. Many cone BC classes also exhibit cross-class coupling with other specific cone BC classes, but only within their ON or OFF superclasses. Such coupling may smooth transitions across BC operating ranges.
Conclusions: Connectomics strategies uniquely afford simultaneous identification of gap junctions and coupling partners in highly complex networks, enabling discovery of novel coupling motifs, target specificities, and spatial correlations. Incorporating coupling topologies and weights into the next generation of network models will provide more powerful predictions of network function.
Funding: NIH/NEI (EY002576, EY015128, EY014800, 1T32EY024234), NSF 0941717, RPB.
Nice work and beautifully illustrated Bryan. Please clarify for me what is meant by weights in the last sentence.