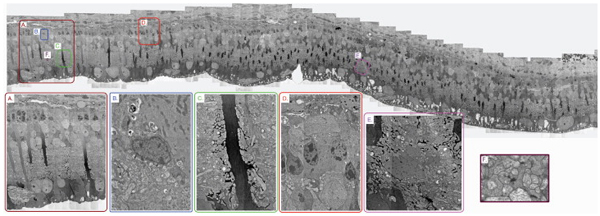
We published an article on the mosaicking and volume assembly of electron microscopy images. This codebase was largely built by Pavel Koshevoy under the direction of Tolga Tasdizen for the connectomics project headed up by Robert Marc. We are tremendously excited for the revised toolset being finished by Brad Grimm who is wrapping much of the functionality together in an easy to implement package called IRIS which will allow us to dramatically increase the amount of imagery we can assemble.
This is amazing! Looked at that paper briefly and there’s quite a few interesting aspects (plus references)! Many of my problems would be solved if our Zeiss confocal allowed me to do tile scans with some overlap between individual scans so that I could semi-automatically arrange the tiles later… unfortunately, it won’t. (I’m a postdoc at Stanford, working on the developing cerebral cortex).
In any case, this is an awesome tool. Thanks for sharing this!
Dino
Hey Dino,
The IR tools actually work with light microscopy data as well. But as you noted, they need som eoverlap with which to calculate an FFT. However, if I recall correctly, the Zeiss confocal software should have a function that allows you to specify overlap of tiles.