We presented an set of work that we are really happy with, giving better than light resolution molecular signals on ultrastructure. PDF is here.
Authors: C. B. Watt, J-H Yang, B. W. Jones, M.V. Shaw, and R. E. Marc.
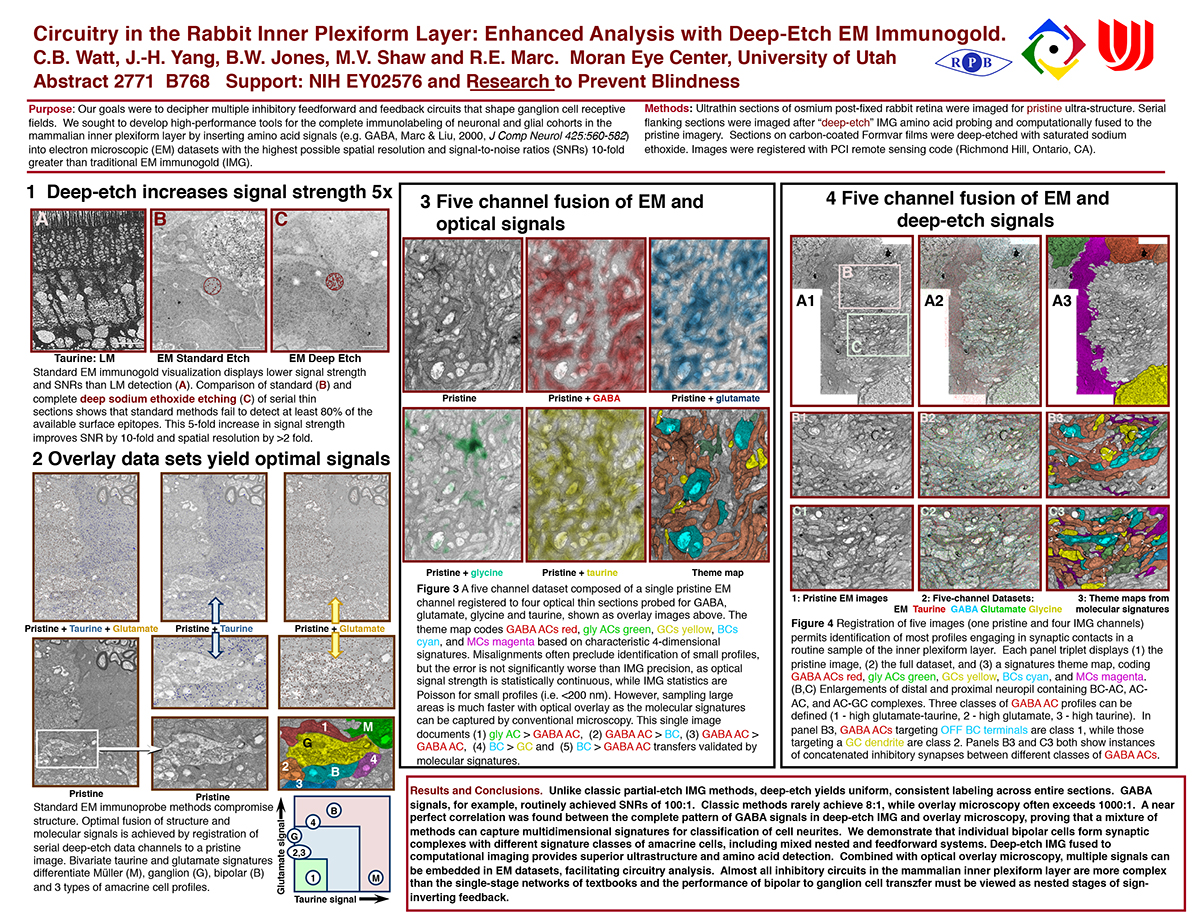
Purpose: Our goals were to decipher multiple inhibitory feedforward and feedback circuits that shape ganglion cell receptive fields. We sought to develop high-performance tools for the complete immunolabeling of neuronal and glial cohorts in the mammalian inner plexiform layer by inserting amino acid signals (e.g. GABA, Marc & Liu, 2000, J Comp Neurol 425:560-582) into electron microscopic (EM) datasets with the highest possible spatial resolution and signal-to-noise ratios (SNRs) 10-fold greater than traditional EM immunogold (IMG).
Methods: Ultrathin sections of osmium post-fixed rabbit retina were imaged for pristine ultra-structure. Serial flanking sections were imaged after “deep-etch” IMG amino acid probing and computationally fused to the pristine imagery. Sections on carbon-coated Formvar films were deep-etched with saturated sodium ethoxide. Images were registered with PCI remote sensing code (Richmond Hill, Ontario, CA).
Results and Conclusions. Unlike classic partial-etch IMG methods, deep-etch yields uniform, consistent labeling across entire sections. GABA signals, for example, routinely achieved SNRs of 100:1. Classic methods rarely achieve 8:1, while overlay microscopy often exceeds 1000:1. A near perfect correlation was found between the complete pattern of GABA signals in deep-etch IMG and overlay microscopy, proving that a mixture of methods can capture multidimensional signatures for classification of cell neurites. We demonstrate that individual bipolar cells form synaptic complexes with different signature classes of amacrine cells, including mixed nested and feedforward systems. Deep-etch IMG fused to computational imaging provides superior ultrastructure and amino acid detection. Combined with optical overlay microscopy, multiple signals can be embedded in EM datasets, facilitating circuitry analysis. Almost all inhibitory circuits in the mammalian inner plexiform layer are more complex than the single-stage networks of textbooks and the performance of bipolar to ganglion cell transzfer must be viewed as nested stages of sign- inverting feedback.